Input format
Clustering and heatmap data are supplied to InCHlib in a Javascript Object Notation (JSON) compliant format. It is an easily extensible text format in which data objects are defined as attribute–value pairs. InCHlib input consists of four blocks (see a color coded example at the end of the page):
- data block - contains heatmap and dendrogram data
- metadata block - contains metadata (i.e., additional information, such as, e.g., class membership, about individual objects)
- column dendrogram block - contains information about a column dendrogram
- column metadata block - contains column metadata (i.e., additional information, such as, e.g., class membership, about individual columns), added in InCHlib 1.1.0
- alternative data block - contains alternative data (i.e., text values that can be displayed instead of original object features), added in InCHlib 1.2.0
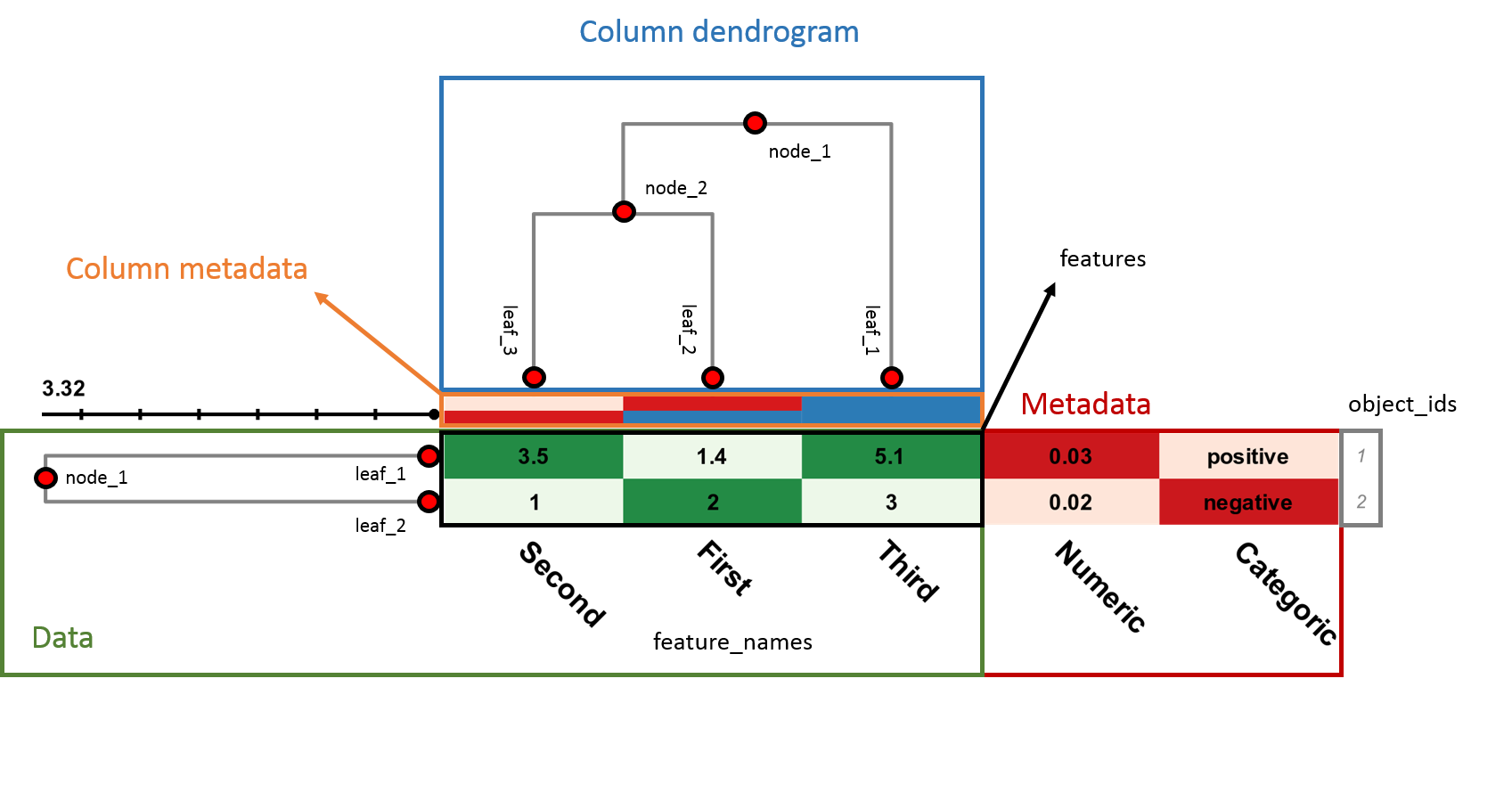
A dendrogram is determined as a structure consisting of inner nodes and terminal nodes (usually referred to as leaves) connected by branches. Each leaf is associated with one data item (row in the heatmap). Each item is described by a set of features (columns in the heatmap). Features describe either one object (data point) or several objects if the row compression is used.
Each node is identified by an arbitrary unique string (ID). While each inner node has two children, no children exist for a leaf. Children of a node are given as left_child and right_child parameters. ID of a parent’s node is given as a parent parameter. The only node without a parent is a root node.
Node example
"node_id": {
"count": 2, //number of items (leafs) which lie in the dendrogram hierarchy below the given node
"distance": 3.32, //distance from the zero base of the dendrogram, given by the distance measure used for clustering
"parent": "node_1", //the ID of a parent node
"left_child": "leaf_1", //ID of a left child
"right_child": "leaf_2" //ID of a right child
},
All leaves line up on the right-hand side of the dendrogram defining the dendrogram zero base. The horizontal axis of the dendrogram measures the distance between clusters. The length of the branch, given as a distance parameter, is measured from the dendrogram zero base utilizing a distance measure used for clustering.
IDs of objects associated with each item (i.e., heatmap row) are given as an objects parameter. Features of each item are stored in a features parameter.
Leaf example
"leaf_id": {
"count": 1, //number of items (leafs) which lie in the dendrogram hierarchy below the given node
"distance": 0, //distance from the zero base of the dendrogram, given by the distance measure used for clustering
"features": [1.4, 3.5, 5.1], //values of individual features defining a data item which is represented by the heatmap row
"parent": "node_1", //the ID of a parent node
"objects": ["object_id"] //list of IDs of objects (data points) represented by a given row
},
Feature_names array defines labels of heatmap columns. Labels for metadata columns are contained under metadata > feature_names. When the column dendrogram is present in the cluster heatmap the column headers are shown under the heatmap, when there is only row dendrogram the labels are above the heatmap.
Feature names example
"feature_names": ["First", "Second", "Third"] //the array of the column feature names
Metadata block contains additional data for data objects, such as, e.g., class membership information. Metadata have no influence on the order of the objects in the heatmap.
Metadata example
"metadata": { //contains nodes and feature_names section of metadata
"feature_names": ["Numeric", "Categoric"], //the array of the metadata feature_names
"nodes": { //contains object IDs with metadata features
"leaf_1": [0.03, "positive"], //the array of metadata features
"leaf_2": [0.02, "negative"]
}
},
Column dendrogram represents the vertical dendrogram and has the same structure as the main row dendrogram contained in nodes section. The only difference is that the terminal nodes (leaves) don't have the features and objects parameters.
Column metadata block contains additional data for data features (columns), such as, e.g., class membership information. Column metadata have no influence on the order of the objects in the heatmap.
Column metadata example
"column_metadata": { //contains features and feature_names sections of column metadata
"features": [ //the array containing arrays of column metadata features
["2", "1", "3"], //the array of column metadata
["negative", "positive", "positive"]
],
"feature_names": ["Numeric", "Categoric"] //the array of the column metadata feature_names
},
Alternative data block contains text values that can be displayed in the heatmap instead of the original object features, e.g., when object features are normalized values, but you want to display raw values (the heatmap coloring is still based on the original object features).
Alternative data example
"alternative_data": { //contains nodes and feature_names section of alt. data
"feature_names": ["Text value 1", "Text value 2", "Text value 3"], //the array of the alt. data feature_names
"nodes": { //contains object IDs with alt. data values
"leaf_1": ["whatever 1", "whatever 2", "whatever 3"], //the array of alt. data values
"leaf_2": ["whatever 1", "whatever 2", "whatever 3"],
}
},
For the generation of InCHlib data format you can use our python wrapper called inchlib_clust.
{
"data": { //contains nodes and feature names section of clustered data
"nodes": { //contains nodes and leafs of row dendrogram
"leaf_1": {
"count": 1, //number of items (leafs) which lie in the dendrogram hierarchy below the given node
"distance": 0, //distance from the zero base of the dendrogram, given by the distance measure used for clustering
"features": [1.4, 3.5, 5.1], //values of individual features defining a data item which is represented by the heatmap row
"parent": "node_1", //the ID of a parent node
"objects": ["object_id"] //list of IDs of objects (data points) represented by a given row
},
"node_1": {
"count": 2,
"distance": 3.32,
"left_child": "leaf_1", //ID of a left child
"right_child": "leaf_2" //ID of a right child
},
"leaf_2": {
"count": 1,
"distance": 0,
"features": [2.0, 1.0, 3.0],
"parent": "node_1",
"objects": ["object_id"]
}
},
"feature_names": ["First", "Second", "Third"] //the array of the column feature names
},
"metadata": { //contains nodes and feature_names section of metadata
"feature_names": ["Numeric", "Categoric"], //the array of the column feature_names
"nodes": { //contains row IDs with metadata values
"leaf_1": [0.03, "positive"], //the array of metadata values
"leaf_2": [0.02, "negative"]
}
},
"column_dendrogram": { //contains nodes section of column dendrogram
"nodes": { //contains nodes of column dendrogram
"leaf_3": {
"count": 1,
"distance": 0,
"parent": "node_2"
},
"node_2": {
"count": 2,
"distance": 2.326,
"left_child": "leaf_2",
"parent": "node_1",
"right_child": "leaf_3"
},
"leaf_1": {
"count": 1,
"distance": 0,
"parent": "node_1"
},
"leaf_2": {
"count": 1,
"distance": 0,
"parent": "node_2"
},
"node_1": {
"count": 3,
"distance": 3.516,
"left_child": "leaf_1",
"right_child": "node_2"
}
}
},
"column_metadata": { //contains features and feature_names sections of column metadata
"features": [ //the array containing arrays of column metadata features
["2", "1", "3"], //the array of column metadata
["negative", "positive", "positive"]
],
"feature_names": ["Numeric", "Categoric"] //the array of the column metadata feature_names
},
"alternative_data": { //contains nodes and feature_names section of alt. data
"feature_names": ["Text value 1", "Text value 2", "Text value 3"], //the array of the alt. data feature_names
"nodes": { //contains object IDs with alt. data values
"leaf_1": ["whatever 1", "whatever 2", "whatever 3"], //the array of alt. data values
"leaf_2": ["whatever 1", "whatever 2", "whatever 3"],
}
}
}
