This use case demonstrates a new feature of InCHlib 1.2 to display images in place of the original heatmap values.
Estrogen alpha receptor (ERα) belongs to the family of nuclear receptors, subfamily of steroid hormone receptors. ERα is a ligand-inducible transcription factor that controls essential physiological, developmental, reproductive and metabolic processes. Estrogen receptors are overexpressed in around 70% of breast cancer cases and have also been implicated in ovarian, colon and prostate cancers. Thus, ERs represent an important target for therapeutic intervention.
From the set of ERα ligands we filtered only these containing steran core structure (the core structure of steroid compounds) with an aromatic A ring (see the picture).
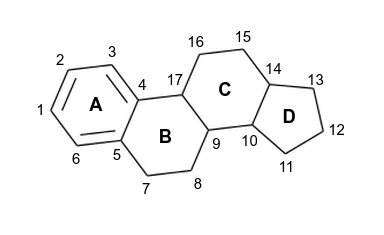
The idea is that if the ligands share the same core the differences in their bioactivities must result from the fragments on individual core positions (Free-Wilson analysis).
Clustered data consist of 1792 bit long binary fingerprints (ECFP4, 256 bits for each fragment). The fingerprints were clustered using Jaccard's distance and Ward's linkage. Since binary data this long are not well suited for a visualization, actual fragments on given positions are displayed instead of the heatmap. pIC50 value (-log(IC50), IC50 = inhibition concentration, i.e., concentration, for which 50% of receptors are inhibitied) is used as a metadata column.
- instances: 22 ERα ligands with steran core structure
- features: molecular fragments on 7 substituted positions (originally clustered by ECFP4 fingerprints, 256 bits for each fragment)
- metadata: pIC50 values of the ligands
When the cursor hovers over the heatmap, ligand structures are shown (the steran core structure is in red). Upon clicking, the heatmap row gets fixed and clicking the depiction of ligand structure on the right opens the corresponding ChEMBL record in a new tab.
