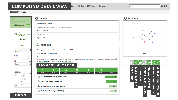
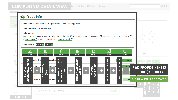
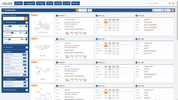
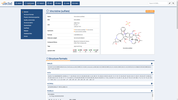
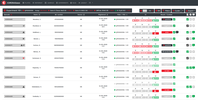
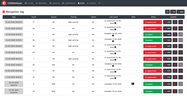
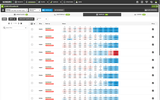
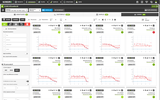
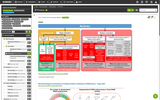
The Probes & Drugs portal (P&D) is a hub for the integration of high-quality bioactive compound sets enabling their analysis and comparison. Its main focus is on chemical probes and drugs but it also includes additional relevant sets from specialist databases/scientific publications, and vendor sets. Upon these, established bioactive chemistry sources (such as ChEMBL, BindingDB, Guide To Pharmacology, DrugCentral or DrugBank) are utilized for compounds' biological annotation. As of ver. 04.2021, the P&D compound database is composed of 76 sources including 14 probe-related, 8 drug compilations, 37 academic/non-commercial sets and 15 precompiled sets from bioactive compound vendors.

HOW TO CITE
Skuta C, Popr M, Muller T, Jindrich J, Kahle M, Sedlak D, Svozil D, Bartunek P. (2017) Probes & Drugs Portal: an interactive, open data resource for chemical biology. Nature Methods 14(8), pp.759-760. http://dx.doi.org/10.1038/nmeth.4365